Weak selection and codon bias evolution in Drosophila
Synonymous, or "silent", DNA changes were considered good candidates for neutral evolution because they do not alter the amino acid sequence of encoded proteins. However, a number of species show preferential usage of codons recognized by abundant tRNA's. This pattern suggests "major codon preference"; natural selection may discriminate among synonymous codons to enhance the speed or the accuracy of protein synthesis (Figure 1).
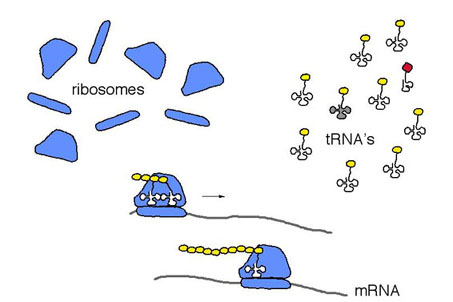
Figure 1. Schematic depiction of protein synthesis.
Under major codon preference, codon usage and tRNA pools are co-adapted to enhance the efficiency of translation. During protein synthesis, the translational apparatus waits at a given codon for the appropriate tRNA to arrive, then links the (yellow) amino acid to the elongating polypeptide. The waiting time is shorter at major codons which are recognized by abundant (unshaded) tRNAs. Shorter waiting times increase the rate of elongation of the polypeptide and decrease the probability that an incorrect amino acid will be incorporated into the protein.
The fitness effects of mutations can be determined by examining patterns of evolution on different time scales. We have developed a statistical method that examines both the frequency spectrum of polymorphisms within populations and the numbers of mutations fixed between species. Computer simulations show that comparisons of such patterns provide a powerful approach to detect even very weak selection (Figure 2). Under major codon preference, silent DNA mutations fall into two fitness classes: "preferred" mutations that increase the number of major codons and "unpreferred" changes in the opposite direction. Frequency distribution and divergence patterns for silent changes in Drosophila simulans are strikingly similar to those predicted by major codon preference; preferred mutations are found at higher frequencies within populations and are more often fixed between species than unpreferred changes (Figure 3). Maximum likelihood estimates support that selection acts at its limit of efficacy in the face of stochastic processes to maintain codon bias in this species.
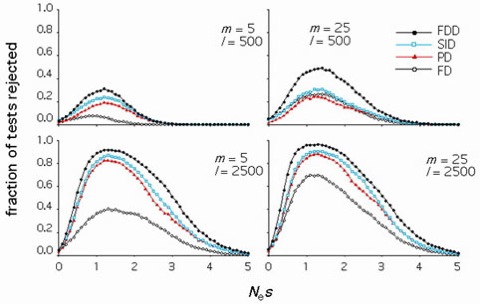
Figure 2. Statistical power to detect weak selection.
The proportion of statistical tests which reject fitness equivalence between preferred and unpreferred mutations simulated under major codon preference are plotted on the y-axis. FD refers to frequency distribution, PD to polymorphism and divergence, SID to singletons, intermediate frequency, and divergence, and FDD to frequency distribution and divergence tests. Data are show for sample sizes of l sites in m alleles.
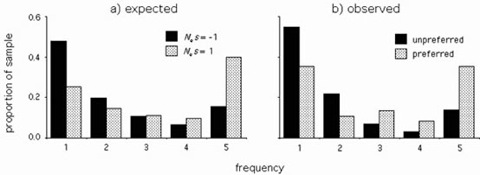
Figure 3. Frequency distributions and divergence of nearly neutral mutations.
a) Expected frequency distributions and divergence under weak selection. For a sample of m = 5 alleles from a population, the expected proportionof mutations found in 1 to 5 of the sequences. b) Observed frequency distributions and divergence of preferred and unpreferred silent DNA mutations among 5 alleles of each of 8 D. simulans genes. The difference between the distributions is significant (Mann-Whitney U-test, P < 0.001).