Vectorette PCR methods
Vectorette PCR was designed for amplification of DNA regions adjacent to known regions and has been employed to amplify 5' and 3' ends of exons as well as long introns (Riley et al. 1990; Arnold and Hodgson 1991). Performing vectorette PCR with multiple vectorette libraries can efficiently obtain orthologous sequences from multiple closely related species. It is useful in molecular evolutionary and systematics studies that require sequences from multiple genes from a group of species.
A DNA vectorette library is constructed by digesting the genomic DNA with a restriction enzyme and followed by the ligation of synthetic oligonucleotides (vectorettes)(Figure 1). PCR amplifications were carried out with one specific primer annealing to the target locus and a vectorette primer priming to the complementary template of vectorette sequence which can only be synthesized after the initial cycle of PCR carried out by the specific primer (Figure 2). The vectorette primer annealing to the complementary template of vectorette sequence is to increase the priming specificity.
Vectorette Library construction, and Vectorette PCR with multiple vectorette libraries
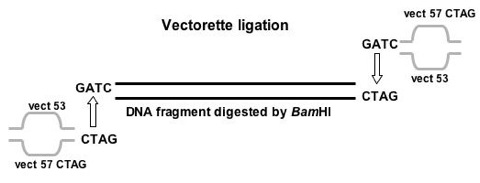
Figure 1. Ligation of vectorette to digested genomic DNA.
An example of vectorette ligation shows that vectorette matches the overhang created by BamHI of a digested DNA fragment. The bubble in the middle of a vectorette represents the mismatched region where PCR primers were designed from this region.
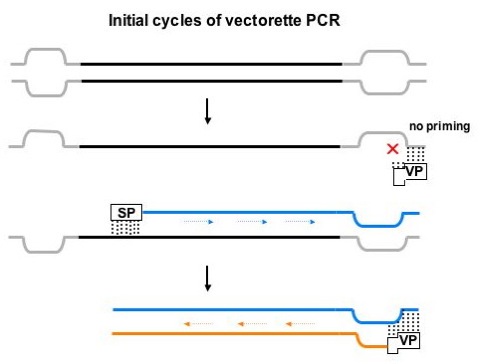
Figure 2. Initial cycles of vectorette PCR.
Vectorette PCR. In the first round of amplification, primer extension only proceeds from the specific primer (SP). Amplification from vectorettes (in gray) does not occur because the vectorette primer (VP) only anneals to the complement of the bottom strand of the vectorette. In the second and subsequent rounds of PCR, priming occurs from both the specific primer and the vectorette primer.
The results of vectorette PCR and following sequencing reaction have confirmed that this method is useful to efficiently obtain sequence information from multiple loci in a group of closely related species. Figure 3 demonstrates one result of vectorette PCR applying to multiple vectorette libraries, which were constructed with a different restriction enzyme in each library. Nine PCR fragments (different sizes in most of them) were successfully amplified in one set of PCR reaction by using one specific primer targeted to the end of Adh exon 3 and a vectorette primer annealing to the vectorette region. A primer designed to anneal to vectorette sequences can apply to these PCR fragments in following sequencing reactions to obtain multiple overlapping sequences in the Adhr region. Since the vectorette primer is universal to all amplified vectorette PCR fragments for all species, this method may reduce the number of primers used for a given locus especially when sequences from multiple species are desired.
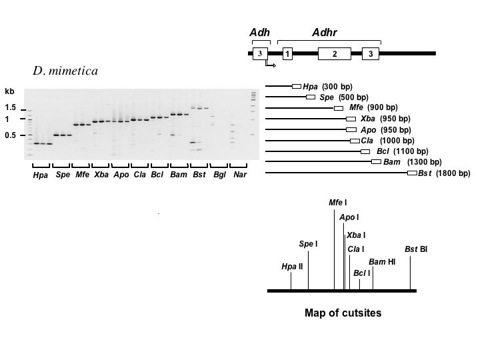
Figure 3. Vectorette PCR of the Alcohol dehydrogenase related (Adhr) gene region from multiple vectorette libraries of Drosophila mimetica.
Gel images show vectorette PCR fragments from amplifications using a specific primer that recognizes the end of the last exon of Adh. Amplifications were performed at three different annealing temperatures (shown from left to right for annealing temperatures from low to high for each vectorette library). The ratio of target to spurious fragments increases with annealing temperature (see BamHI and BstBI lanes in the figure above). Size standards are shown in the right- and left-most lanes of each gel. Vectorette PCR products are shown in increasing order of size and the corresponding regions of the Adhr region are depicted to the right of each gel image. The empty box at the right end of each PCR product represents the vectorette sequence. Results are also shown for two libraries that were not expected to yield fragments (Bgl II and Nar I). The map of restriction enzyme cut sites from the known sequence of Adhr in D. mimetica is also shown below those PCR fragments. All expected fragments were successfully amplified.
References
- Molecular phylogeny of the Drosophila melanogaster species subgroup
- J Mol Evol 57:562–573. 2003. supplementary material
- Vectorette PCR: a novel approach to genomic walking
- PCR Methods Appl 1:39-42. 1991
- A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones.
- Nucleic Acids Res 18:2887-90. 1990