Phenotypic basis of selection at silent sites: Codon bias and translational accuracy
Population genetic studies of DNA data are often limited by a lack of knowledge of the phenotypes on which natural selection acts. At silent sites, however, the functional basis of fitness variation can be determined from comparative DNA sequence analyses.
Synonymous codon usage may affect the fidelity of protein synthesis. Because the fitness cost of a translational misincorporation depends on how severely it disrupts protein function, selection for translational accuracy predicts an association between codon usage in DNA and functional constraint at the protein level. Constrained amino acid positions can be identified either by evolutionary conservation or by functional assays. In D. melanogaster, constrained amino acid positions show higher major codon usage than less constrained positions. Natural selection appears to act at silent sites to enhance the accuracy of protein synthesis.
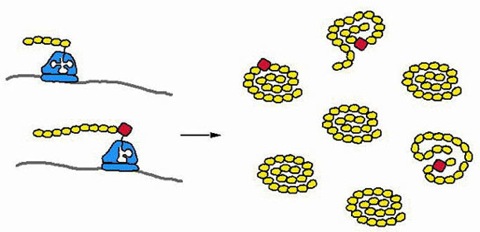
Figure 1. Errors in protein synthesis.
mRNAs are translated into a population of proteins (on the right). The homopolymer of yellow amino acids is the protein sequence encoded in the gene. However, because about 1 in 1000 translation events results in an amino acid misincorporation, a large fraction of the proteins will contain non-coded amino acids. The functional effect of such misincorporations will vary among positions in the protein.
References
- Synonymous Codon Usage in Drosophila Melanogaster: Natural Selection and Translational Accuracy
- Genetics 136:927–935. 1994.