Molecular phylogeny of the Drosophila melanogaster species subgroup
The Drosophila melanogaster species subgroup consists of nine species that appear to be of Afro-tropical origin. The human commensals, D. melanogaster and D. simulans, are cosmopolitan in their distributions. D. sechellia and D. mauritiana are endemic island species closely related to D. simulans. D. teissieri and D. yakuba have similar geographic ranges spreading from northwest to southeast Africa. D. teissieri is mainly a forest species while D. yakuba, is more frequently found in a savanna habitat. D. erecta and D. orena are restricted to west central Africa (Lachaise et al. 1988). Finally, D. santomea, a close relative of D. yakuba, was recently discovered on Sao Tome island in the Gulf of Guinea in West-equatorial Africa (Lachaise et al. 2000). Although molecular and phenotypic evolution have been studied extensively in D. melanogaster and its close relatives, phylogenetic relationships within the D. melanogaster species subgroup remain unresolved. We reconstruct the phylogeny of the melanogaster species subgroup using DNA sequence data from four nuclear genes (totaling 7164 bp) from six melanogaster subgroup species (D. melanogaster, D. simulans, D. teissieri, D. yakuba, D. erecta, and D. orena) and three species from subgroups outside the D. melanogaster species subgroup [D. eugracilis (eugracilis subgroup), D. mimetica (suzukii subgroup), and D. lutescens (takahashii subgroup)]. Phylogenetic analyses strongly support a D. yakuba-D. teissieri and D. erecta-D. orena clade within the D. melanogaster species subgroup. D. eugracilis is grouped closer to the D. melanogaster subgroup than a D. mimetica-D. lutescens clade.
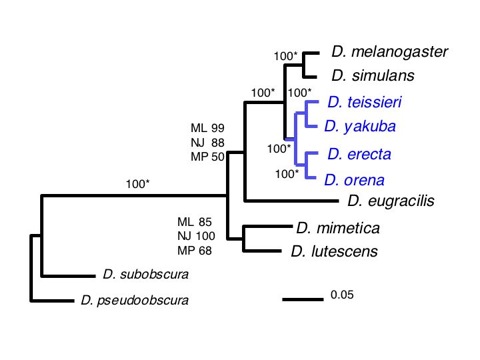
Figure 1. Maximum likelihood trees of species in the melanogaster group inferred from a concatenated sequence of Adh + Adhr + Gld + ry genes (7164 bp).
A Bootstrap consensus tree. Bootstrap values (1000 replicates) on each node are shown and represent, from top to bottom: Maximum likelihood(ML), neighbor-joining distance (NJ), and maximum parsimony (MP) values. “ * ” indicates identical bootstrap values for ML, NJ, and MP.
References
- Molecular phylogeny of the Drosophila melanogaster species subgroup
- J Mol Evol 57:562–573. 2003. supplementary material
- Historical Biogeography of the Drosophila melanogaster Species Subgroup
- Evolutionary Biology 22:159-225. 1988
- Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tome
- Proc R Soc Lond B Biol Sci 267:1487-95. 2000