Mechanisms of protein evolution
Selection at silent sites has important implications for identifying the forces governing the evolution of other classes of molecular change. Several claims of adaptive protein evolution rely on comparisons between the dynamics of synonymous and replacement mutations. These arguments depend critically on an assumption of neutrality at silent sites; major codon preference provides an alternative explanation for a number of these findings.
Selection at silent sites can, however, be employed to shed light on mechanisms of protein evolution. D. melanogaster has undergone a genome-wide reduction in the efficacy of selection for codon bias; intron base composition has remained constant while codon bias has undergone a dramatic and genome-wide decline. Accelerated rates of amino acid substitution and increases in protein sizes in the D. melanogaster lineage suggest the fixation of mildly deleterious changes in protein structure. In contrast, in D. simulans, silent DNA evolution appears to reflect a balance among mutation pressure, natural selection and genetic drift. In this lineage, amino acid mutations show an excess of fixed differences and a deficiency of intermediate frequency polymorphisms. This pattern is consistent with a combination of adaptive and weakly deleterious protein evolution and suggests that a surprisingly small fraction of protein changes evolve neutrally (Figure 5).
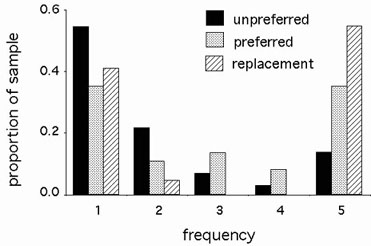
Figure 1. Frequency distributions and divergence of silent and replacement mutations.
Observed FDD patterns among 5 alleles of each of 8 D. simulans genes. The differences between unpreferred and replacement (P < 0.001) and preferred and replacement (P = 0.028) mutations are significant.
References
- Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster.
- Genetics 144:1297–1307. 1996.
- Inferring the fitness effects of DNA mutations from polymorphism and divergence data: statistical power to detect directional selection under stationarity and free recombination
- Genetics 151:221–238. 1999
- Codon Usage Selection Can Bias Estimation of the Fraction of Adaptive Amino Acid Fixations
- Mol Biol Evol 33:1580–1589. 2016.